
The Systems Biology graphical Notation
The Systems Biology Graphical Notation (SBGN) is a standard graphical representation intended to foster the efficient storage, exchange and reuse of information about signaling pathways, metabolic networks, and gene regulatory networks amongst communities of biochemists, biologists, and theoreticians. SBGN is made up of three complementary languages: Process descriptions, Activity flows, and Entity relationships. SBGN has been developed over many years by a diverse community of systems biologists, software developers and knowledge representation specialists. Its grammar is described in very detailed specifications, and is supported by an ecosystem of software tools.
The SBGN languages |


|
The complexity of biological structures and their functions mean that a single type of graphical representation cannot be used to represent every situations. For instance, metabolic pathways are often represented as a series of chemical reactions, where each reactant is represented. However, such a representation explodes when we try to use if with entities with many components or many states (e.g., protein complexes, allosteric proteins, proteins with many postranslational modifications). On the contrary, signalling pathways or genetic regulations are generally represented as influences from one entity onto another. Such a compact representation nevertheless cannot represent the consequence of such influences on their source. SBGN is thus made up of three orthogonal languages, each with its strengths and weaknesses.
 The Process Description language is made up of two different types of nodes (PDs are bipartite graphs) representing pools of physical entities ("continuants") and processes ("occurants", e.g., reactions. The arcs linking the nodes represent consumption, production, and modulation. Nodes can be adorned with states. This language allows a very detailed description of all the processes taking place in a system. However, it is subjected to combinatorial explosion. Process Descriptions are often used in most metabolic networks (e.g. Reactome, KEGG Pathways metabolic pathways, etc.). They are good graphical representations of process modelling, e.g. ODEs or stochastic processes. SBGN PD glyphs are summarised in the reference card:
The Process Description language is made up of two different types of nodes (PDs are bipartite graphs) representing pools of physical entities ("continuants") and processes ("occurants", e.g., reactions. The arcs linking the nodes represent consumption, production, and modulation. Nodes can be adorned with states. This language allows a very detailed description of all the processes taking place in a system. However, it is subjected to combinatorial explosion. Process Descriptions are often used in most metabolic networks (e.g. Reactome, KEGG Pathways metabolic pathways, etc.). They are good graphical representations of process modelling, e.g. ODEs or stochastic processes. SBGN PD glyphs are summarised in the reference card: 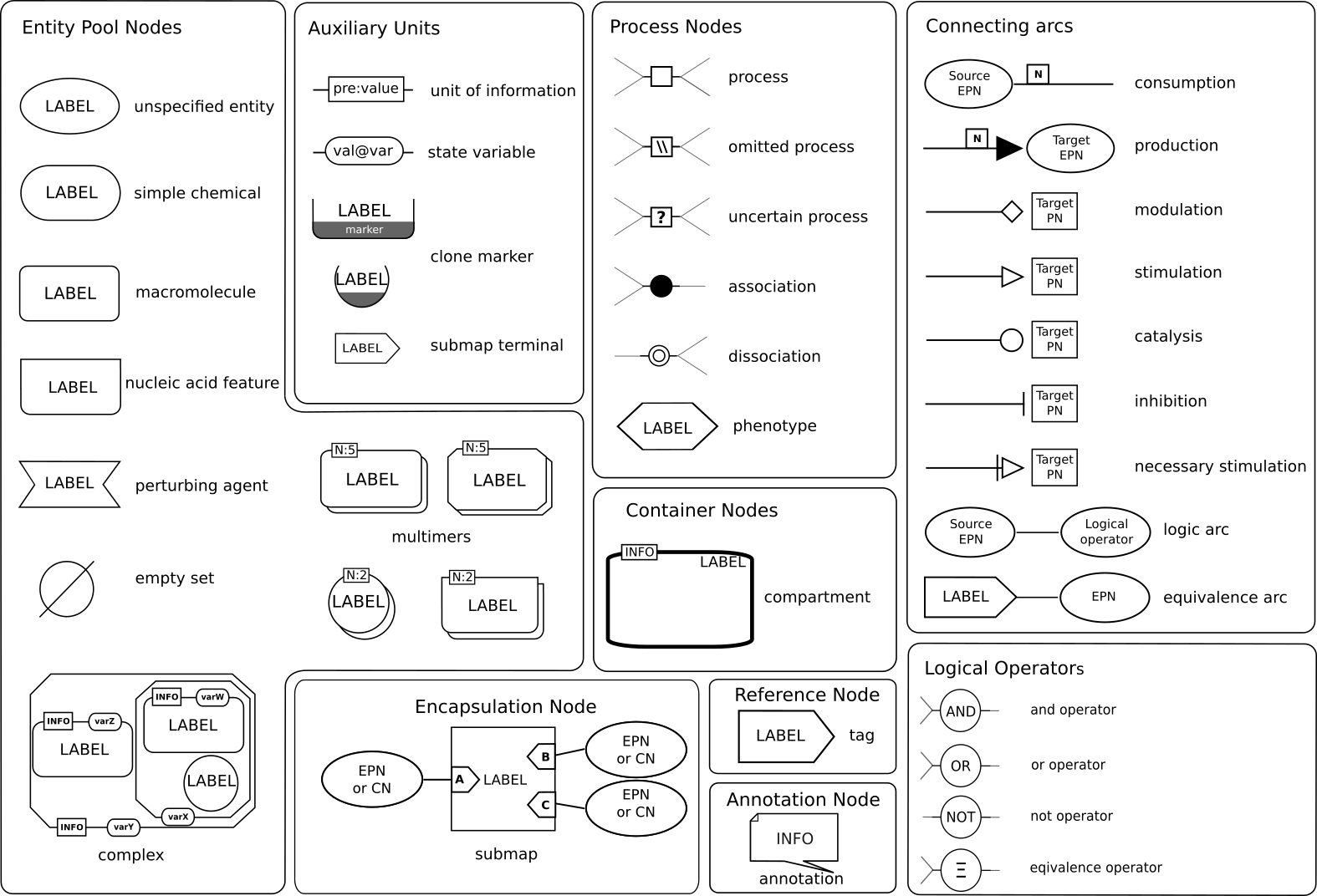 Also available in SVG
Also available in SVG
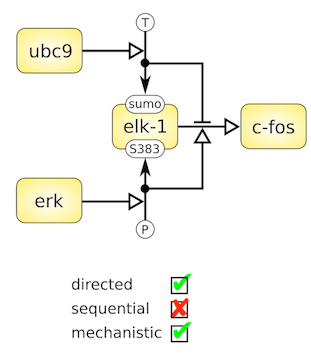
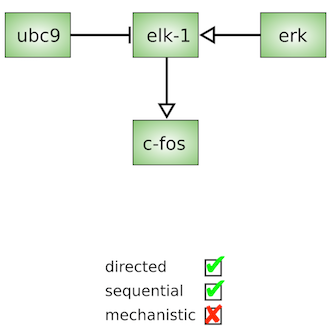 The Activity Flows language is made up of only one type of node representing activities carried by entities (Thus only "occurants"). The arcs linking the nodes represent influences such as stimulations, inhibitions, etc. The language is very compact, but cannot represent the mechanistic details underlying an effect. Activity Flows are often used in signalling pathway databases (e.g. KEGG pathways signalling pathways). Note that SBGN AF is not a compact version of SBGN PD, but a semantically different language. However, in some situations, a conversion PD to AF is possible (Vogt et al. 2013). SBGN AF glyphs are summarised in the reference card:
The Activity Flows language is made up of only one type of node representing activities carried by entities (Thus only "occurants"). The arcs linking the nodes represent influences such as stimulations, inhibitions, etc. The language is very compact, but cannot represent the mechanistic details underlying an effect. Activity Flows are often used in signalling pathway databases (e.g. KEGG pathways signalling pathways). Note that SBGN AF is not a compact version of SBGN PD, but a semantically different language. However, in some situations, a conversion PD to AF is possible (Vogt et al. 2013). SBGN AF glyphs are summarised in the reference card: 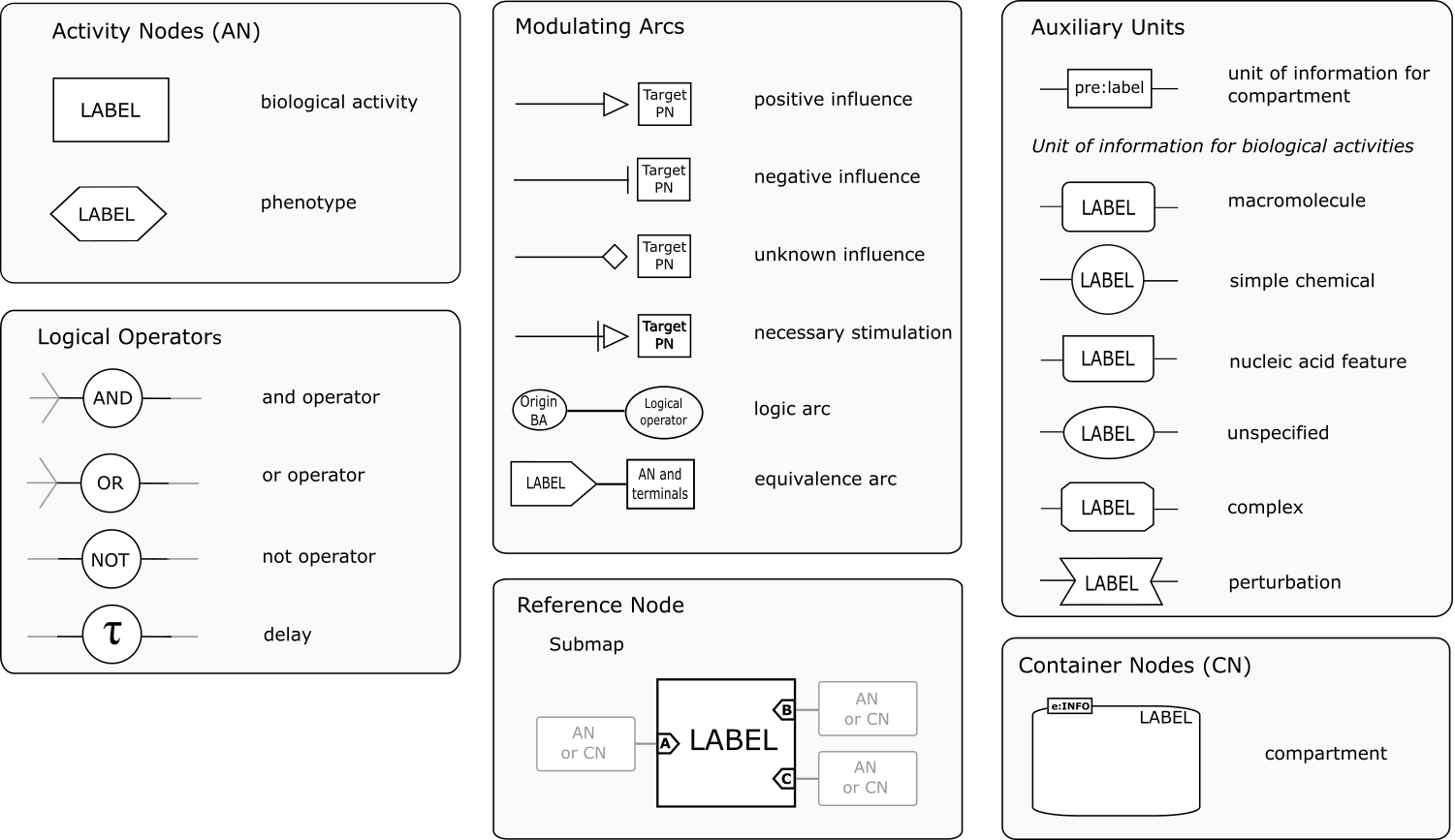 Also available in SVG
Also available in SVG
 The Entity Relationship language is made up of only one type of node representing entities (Thus only "continuants"). The arcs linking the nodes represent relationships, such as modulation or binding. The language is very compact and permit to avoid combinatorial explosion, but cannot represent successions of events. Each relationship is independent.SBGN ER glyphs are summarised in the reference card:
The Entity Relationship language is made up of only one type of node representing entities (Thus only "continuants"). The arcs linking the nodes represent relationships, such as modulation or binding. The language is very compact and permit to avoid combinatorial explosion, but cannot represent successions of events. Each relationship is independent.SBGN ER glyphs are summarised in the reference card: 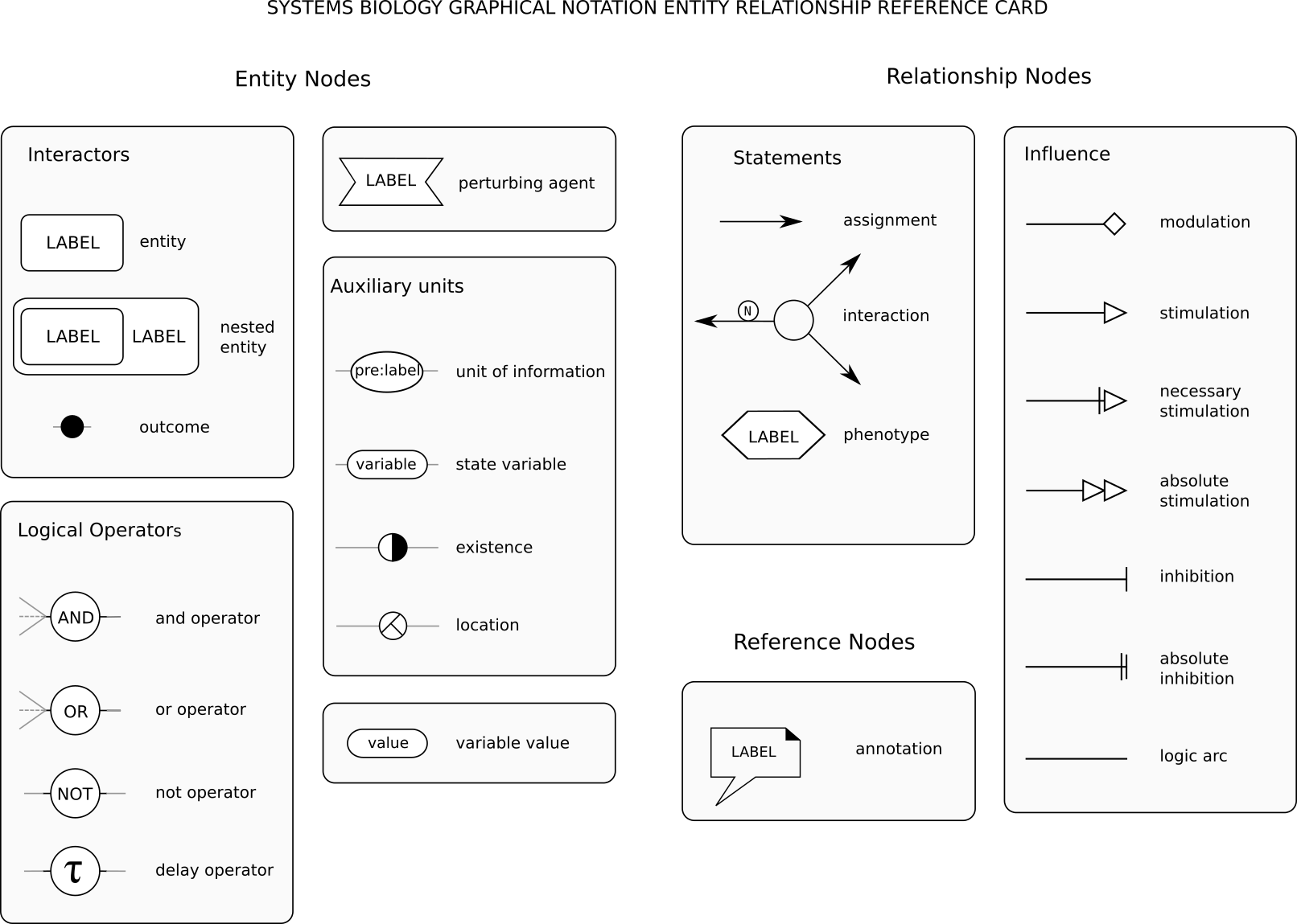 Also available in SVG
Also available in SVG
References |


|
The main paper describing SBGN
The Systems Biology Graphical Notation.
Nature Biotechnology
(2009), 27: 735-741
The Systems Biology Graphical Notation. Nature Biotechnology (2009), 27: 735-741.
[]
[
 ]
[Supplementary info]
]
[Supplementary info]
Latest specifications
Systems Biology Graphical Notation: Process Description language Level 1 Version 2.0.
Journal of Integrative Bioinformatics
(2019) 002
[]
Systems Biology Graphical Notation: Entity Relationship language Level 1 Version 2.
Journal of Integrative Bioinformatics
(2015) 12(2): 264
[]
Systems Biology Graphical Notation: Activity Flow language Level 1 Version 1.2.
Journal of Integrative Bioinformatics
(2015) 12(2): 266
[]
Other publications of interest
Quick tips for creating effective and impactful biological pathways using the Systems Biology Graphical Notation.
PLoS Computational Biology
(2018) 14(2): e1005740
[]
[
 ]
]
Translation of SBGN maps: Process Description to Activity Flow.
BMC Systems Biology
(2013) 7:115
[]
Related links |


|
Examples of nice SBGN diagram from the literature |


|
SBGN diagrams can be both precise and nice-looking. Colors and decorations are not part of the standard and should not convey exchangeable semantics. However, we are free to embellish SBGN diagrams as much as we want or need. Here are a few we can find in the scientific literature (mostly from my papers at the moment, since I did not have to search for them...). Open in a new tab to see them full size.
Quick tips for creating effective and impactful biological pathways using the Systems Biology Graphical Notation.
PLoS Computational Biology
(2018) 14(2): e1005740
[]
[
 ]
]
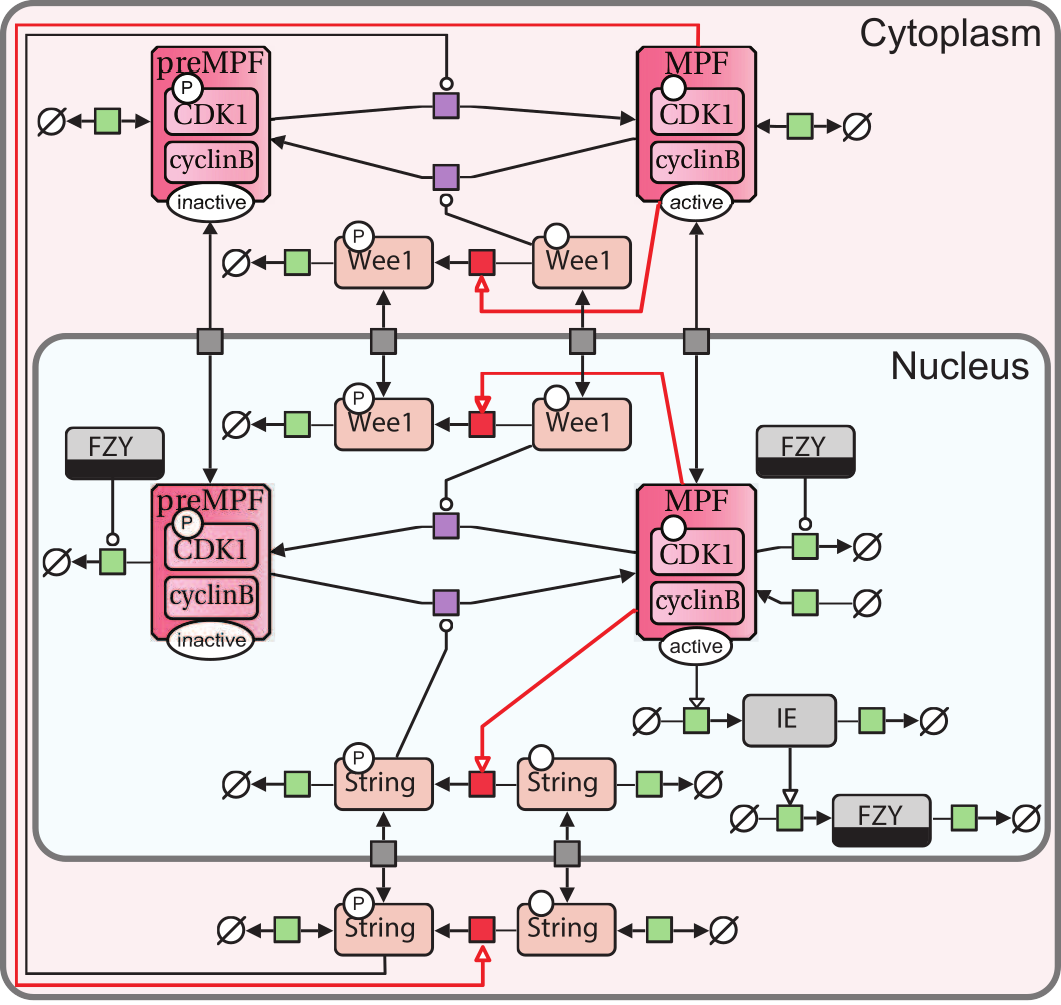 The paper presented the steps to beautify an SBGN map. The image presents Process Descriptions describing the regulation of cell cycle.
The paper presented the steps to beautify an SBGN map. The image presents Process Descriptions describing the regulation of cell cycle.
Reciprocal regulation of ARPP-16 by PKA and MAST-3 kinases provides a cAMP-regulated switch in protein phosphatase 2A inhibition. eLife (2017) 6: e24998
[
 ]
]
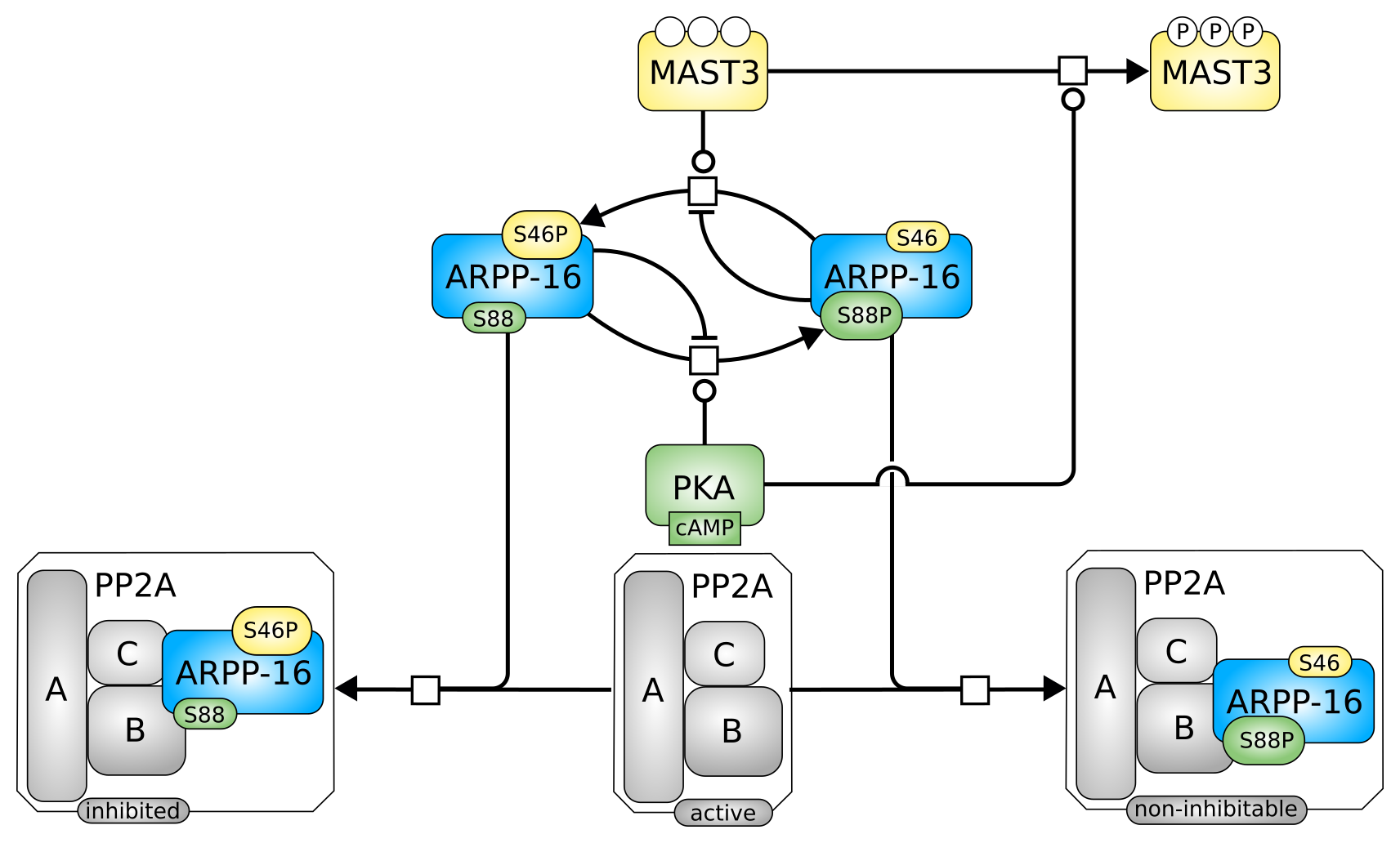 Process Descriptions describing the regulation of ARPP-16 by protein phosphatase 2A.
Process Descriptions describing the regulation of ARPP-16 by protein phosphatase 2A.
Significance of stroma in biology of oral squamous cell carcinoma. Tumori (2017) 104(1): 9-14
[
 ]
]
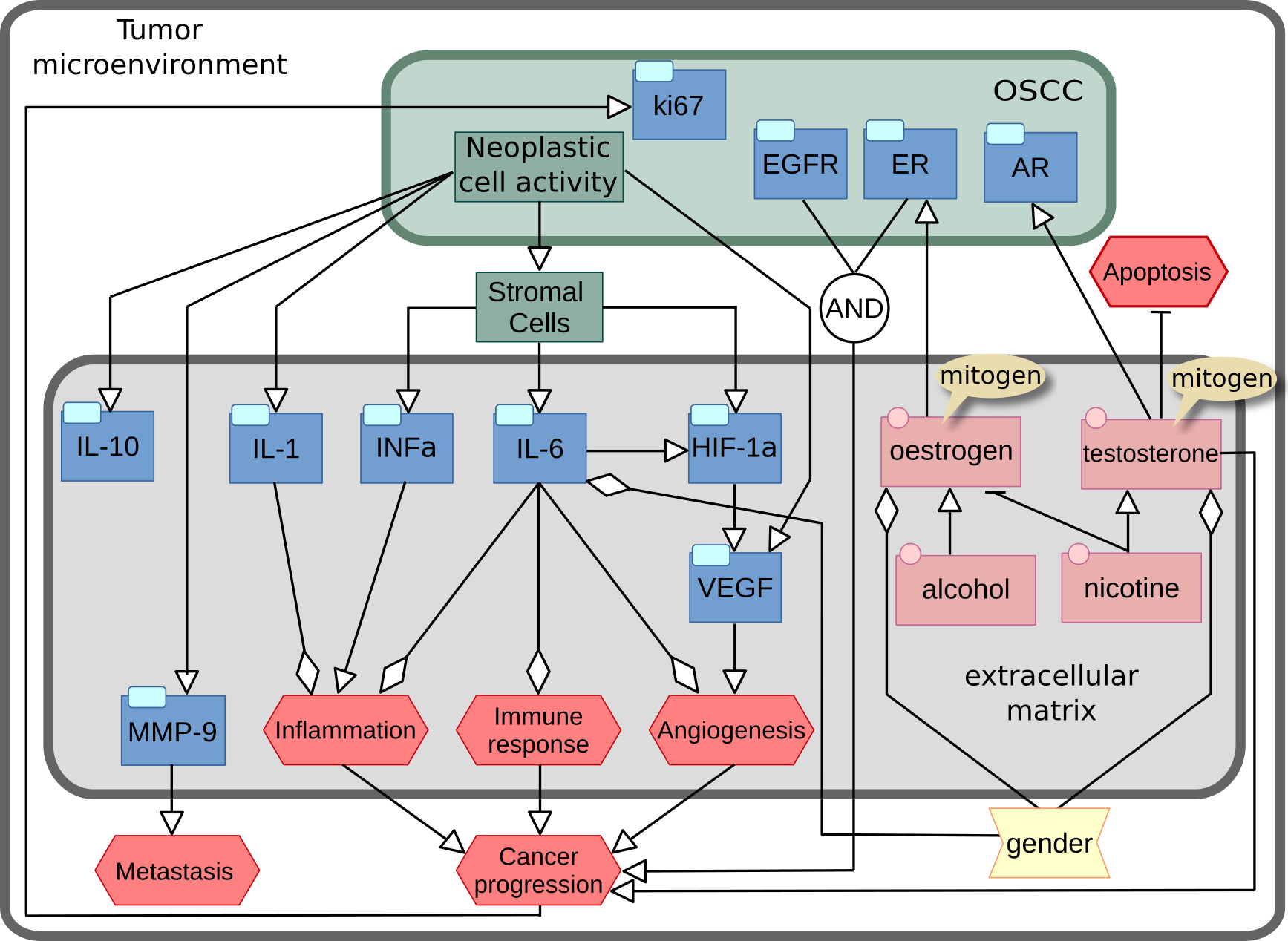
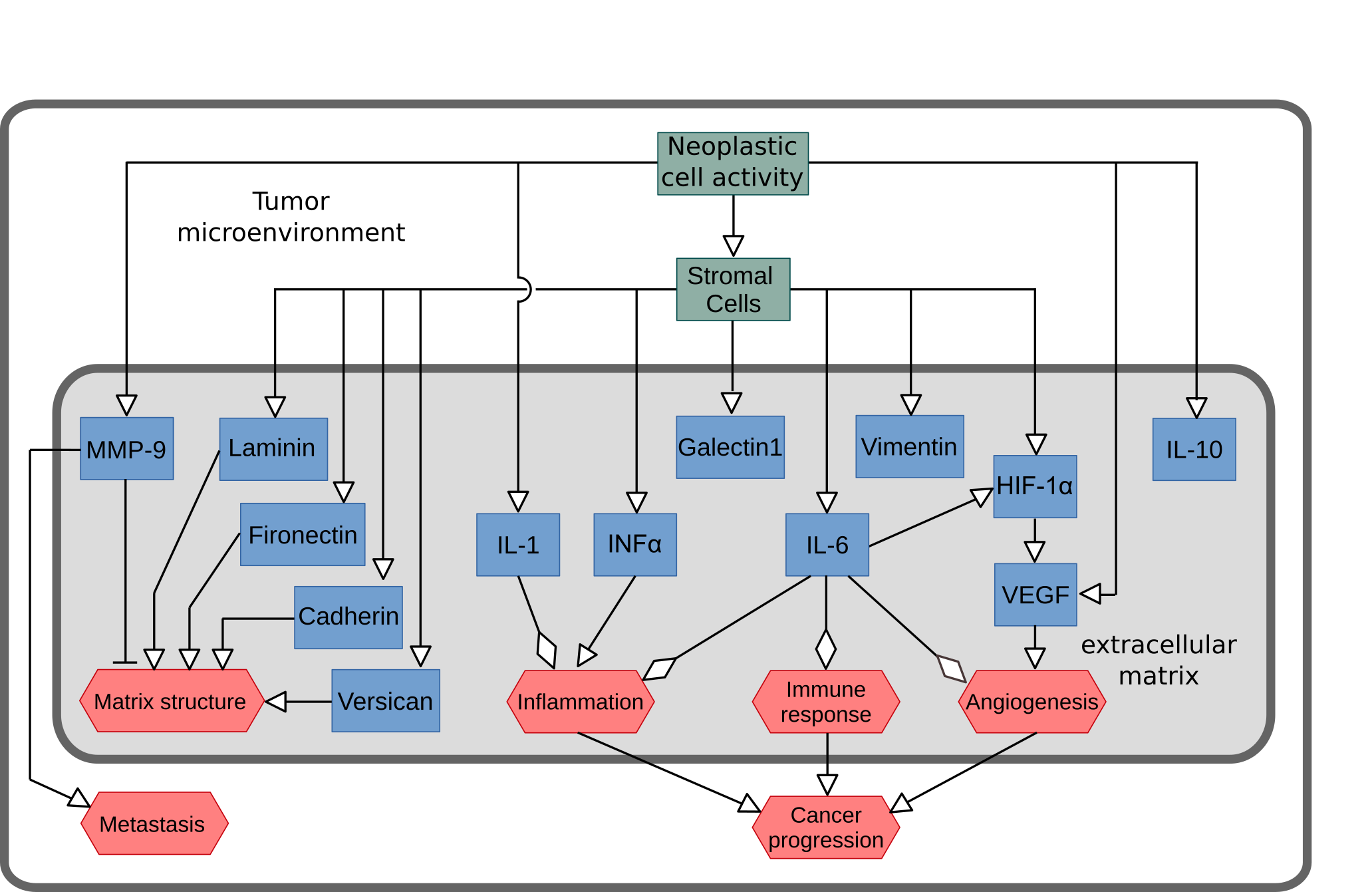 Activity Flows describing the regulation of microenvironment in oral squamous cell carcinoma. Two successive versions were designed, and I preferred the initial, unpublished, one (below)
Activity Flows describing the regulation of microenvironment in oral squamous cell carcinoma. Two successive versions were designed, and I preferred the initial, unpublished, one (below)
The impact of mathematical modeling in understanding the mechanisms underlying neurodegeneration: evolving dimensions and future directions. CPT: Pharmacometrics & Systems Pharmacology (2016), 6:73–86 [
 ]
]
In this review of computational models used in the field of neurodegenerative diseases, we presented two maps.
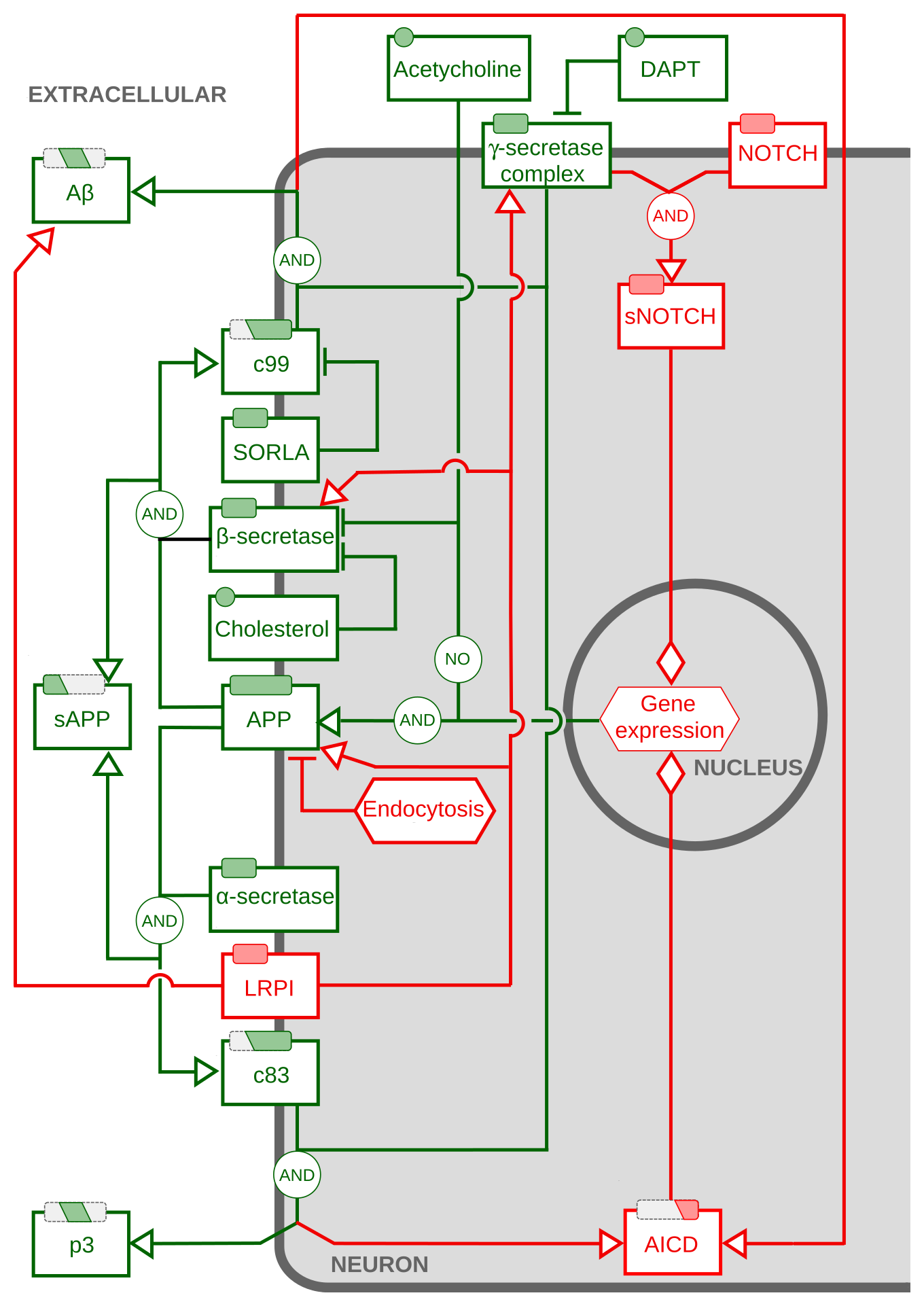 Activity Flows describing the processing of amyloid-beta precursor protein.
Activity Flows describing the processing of amyloid-beta precursor protein.
 Process Descriptions of amyloid-beta aggregation.
Process Descriptions of amyloid-beta aggregation.
Quantitative and logic modelling of gene and molecular networks. Nature Reviews Genetics (2015) 16: 146–158 [
 ]
]
 Process Descriptions of a minimal gene regulatory network.
Process Descriptions of a minimal gene regulatory network.
A community-driven global reconstruction of human metabolism. Nature Biotechnology (2013) 31: 419–425. [
 ]
]
This process description map corresponds to the model presented in the paper, although it was not published with the paper, but distributed via the Virtual Metabolic Human.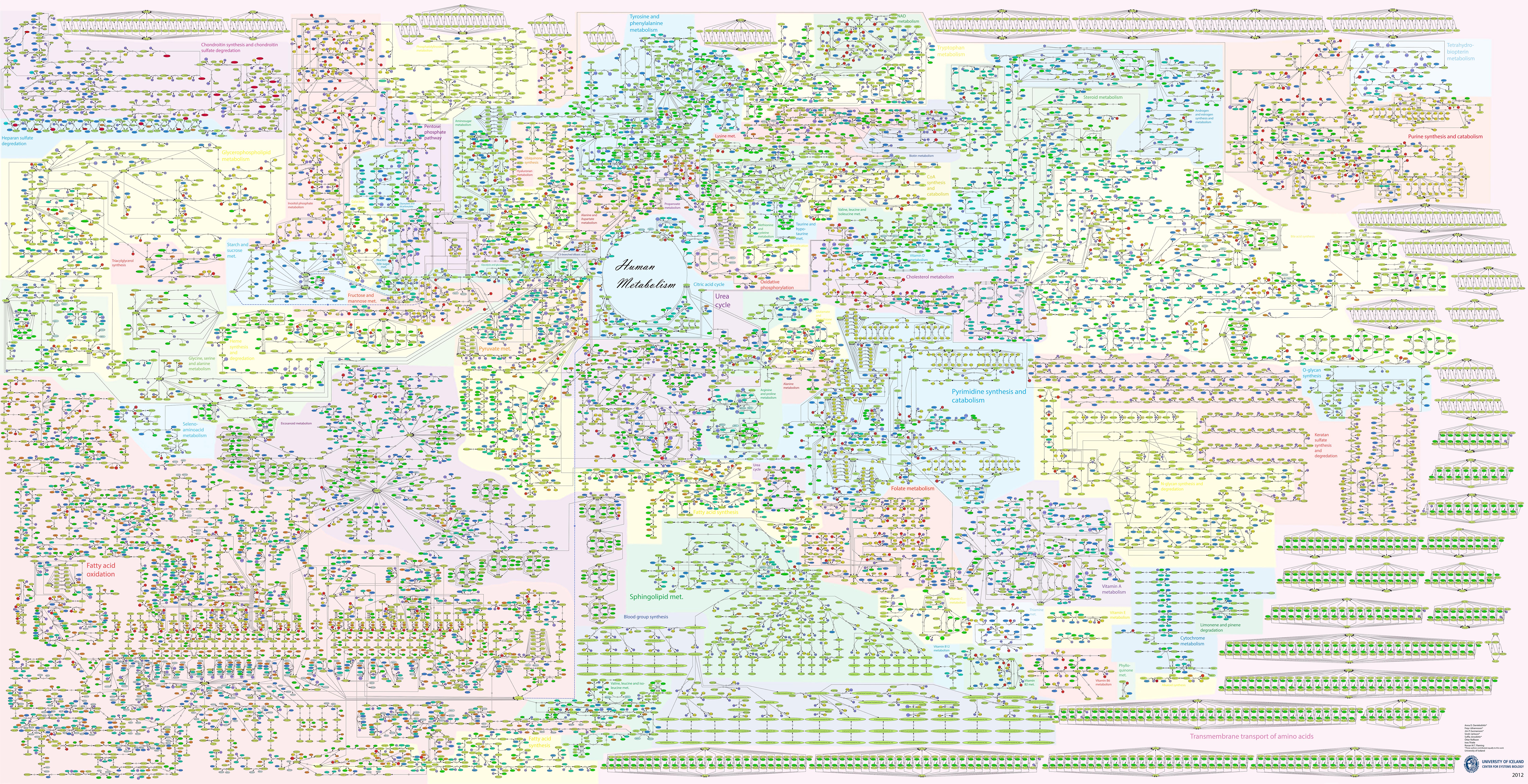
Structural analysis and stochastic modelling suggest a mechanism for calmodulin trapping by CaMKII. PLoS ONE (2012), 7(1): e29406 [
 ]
]
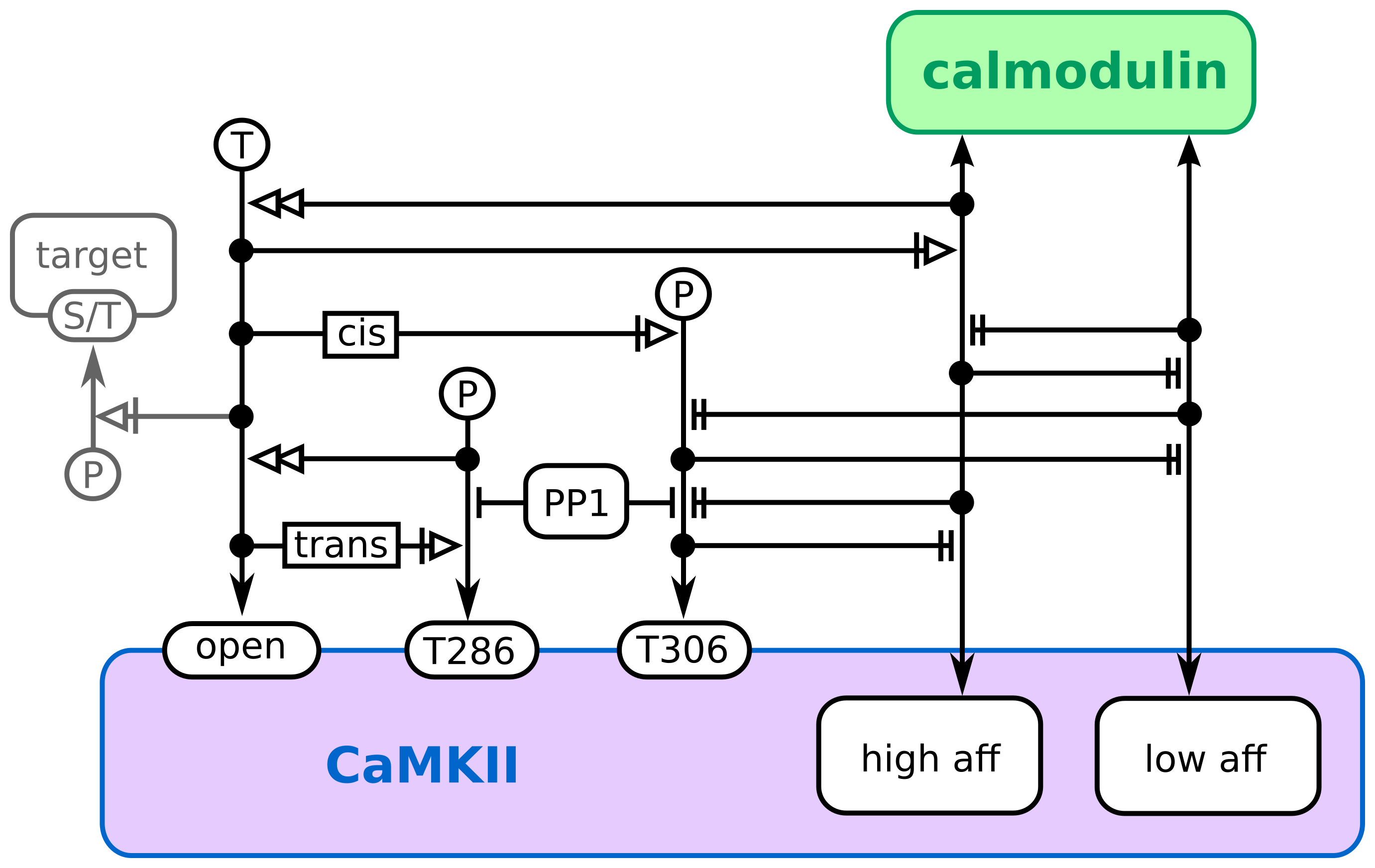 Entity Relationships describing the regulation of calcium/calmodulin kinase II.
Entity Relationships describing the regulation of calcium/calmodulin kinase II.
The Systems Biology Graphical Notation. Nature Biotechnology (2009), 27: 735-741. [
 ]
]
In the main SBGN papers, we presented three examples to illustrate the three languages
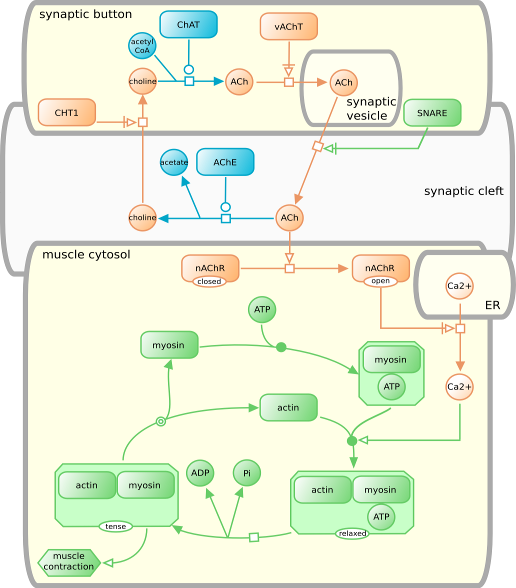 Process Descriptions of muscle cell activation by acetylcholine.
Process Descriptions of muscle cell activation by acetylcholine.
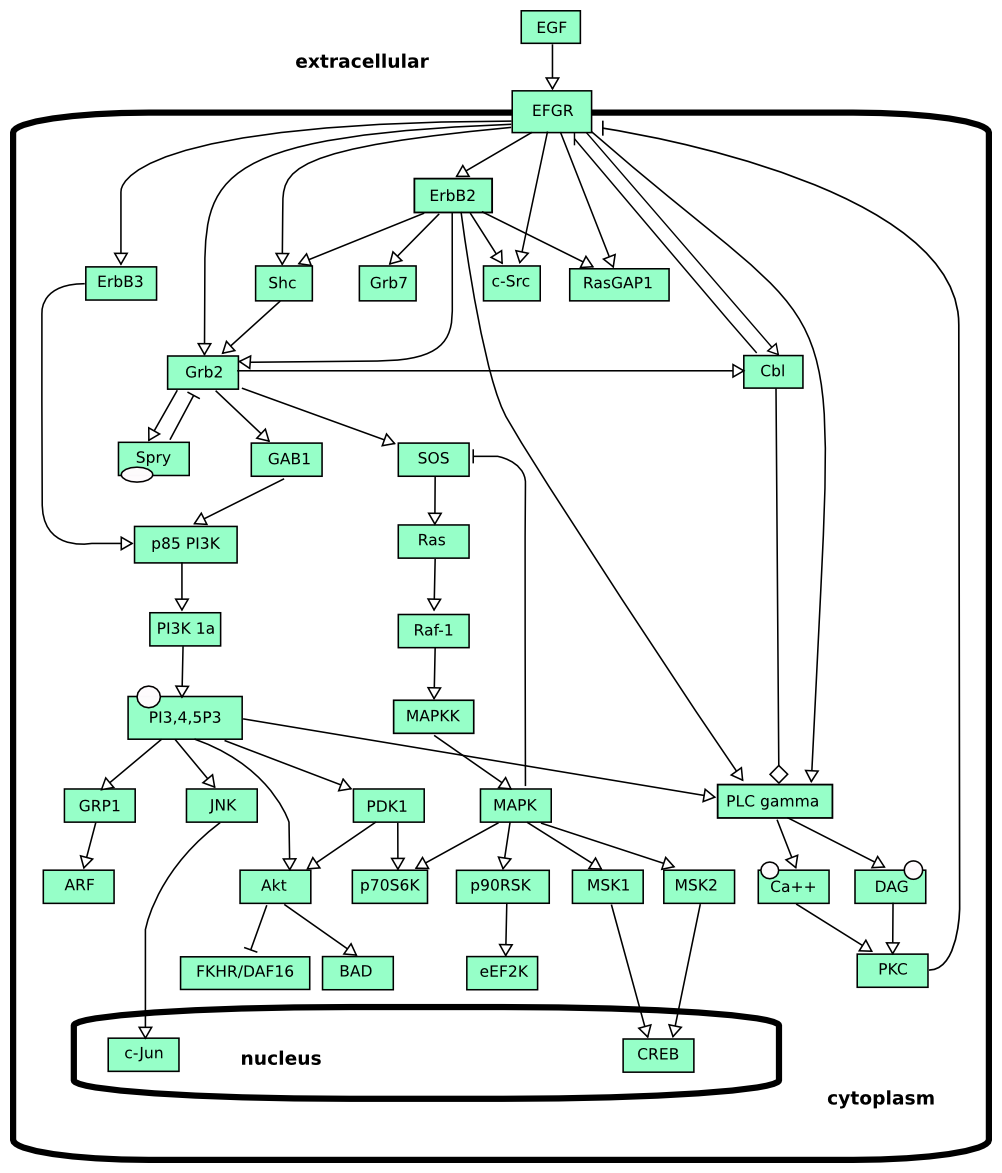 Activity flow of Epidermal growth factor signalling.
Activity flow of Epidermal growth factor signalling.
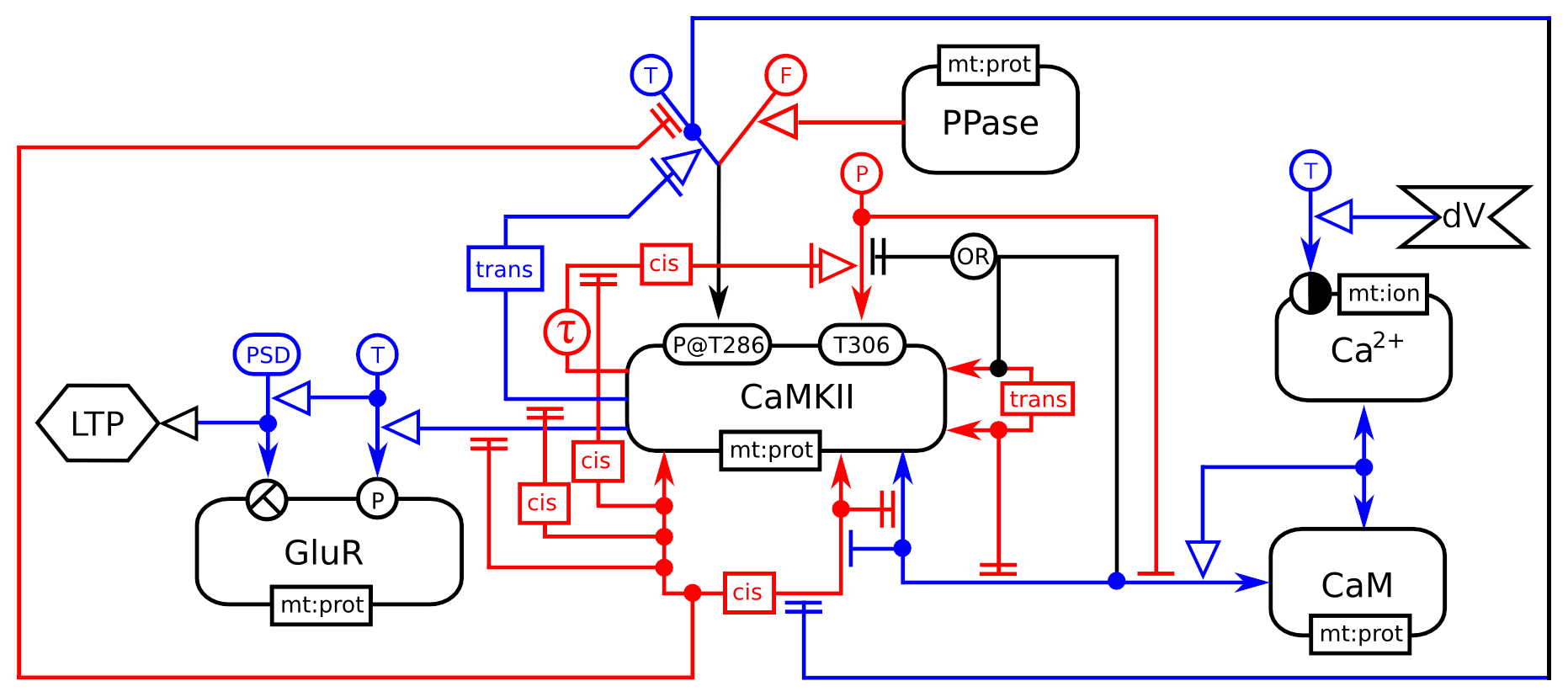 Entity relationship of calcium calmoduline kinase II regulation in synaptic plasticity.
Entity relationship of calcium calmoduline kinase II regulation in synaptic plasticity.